Because of the needs to use libraries and tools in different languages and to execute them on different systems such as computer clusters, bioinformaticians write a lot of scripts in different languages and face many challenges in developing, running, managing, sharing, and reproducing bioinformatic data analyses. Notably,
- Management of scripts: With increasing number of scripts, some in multiple versions, some shared among projects, some written for and executed on remote systems, it can be difficult to share data analyses with others, and reproduce prior data analyses at a later time.
- Analyze large datasets: It is tedious and error-prone to write scripts to analyze larger and larger amount of data as more and more work is needed for the “execution” part of the analysis. Additional work includes but not limited to refactoring scripts for batch execution, creation and submission of jobs, monitoring and fault recovery, and synchronization of data and results across system.
Why workflow systems are not suitable for daily computational research
Scientific pipeline systems are supposed to alleviate these problems by streamlining the creation and execution of data analysis workflows. However, after surveying all 100+ workflow systems and trying more than 10 of them, I realized that the overhead of applying a workflow system in daily data analysis is too high to help productivity. As a matter of fact, as pointed out by Loman and Watson, 2013, Nat Biotechnol, using a workflow system for analyses that will be run only once, or not meant to be shared with or repeated by others can decrease creativity and productivity.
Pipelineitis is a nasty disease
A pipeline is a series of steps, or software tools, run in sequence according to a predefined plan. Pipelines are great for running exactly the same set of steps in a repetitive fashion, and for sharing protocols with others, but they force you into a rigid way of thinking and can decrease creativity.
Warning: don’t pipeline too early. Get a method working before you turn it into a pipeline. And even then, does it need to be a pipeline? Have you saved time? Is your pipeline really of use to others? If those steps are only ever going to be run by you, then a simple script will suffice and any attempts at pipelining will simply waste time. Similarly, if those steps will only ever be run once, just run them once, document the fact you did so and move on.
In another word, workflow systems are designed for repeated execution of mature workflows for which a good deal of work is invested to help the users of workflows. They are not designed for daily computational research where scripts are written for particular projects and are almost disposable.
Pipelineitis is treatable
Pipelineitis is a nasty disease but admitting the facts does not save us from all the aforementioned problems we have in daily data analysis. Looking over and over at the key obstacles that prevent the application of workflow systems in daily computational research, we realized that narrowing the gap between environments for interactive data analysis and workflow systems for batch data crunching would cure pipelineitis and allow the use of workflow systems in daily computational research. With this idea in mind, we developed SoS (Script of Scripts), which is a workflow engine with a multi-language notebook frontend. The following table summarizes the major features of SoS and how they help alleviate the symptoms of pipelineitis:
| Symptom | Cause | Treatment |
|---|---|---|
| Lower productivity | Re-create data analysis using another language or environment |
|
| Steep learnin curve | Workflow language difficult to learn and use, making it difficult to adopt |
|
| Difficult to read and share | Data analysis logics (core steps)
|
|
| Difficult to reproduce | Data analysis scattered in multiple scripts for multiple systems |
|
An IDE for both interactive data analysis and workflow development
A complex workflow inevitably involves scripts in multiple languages. Existing workflow systems generally do not provide an interactive environment for the development of these scripts. Even for workflow systems that provide nice GUIs, the graphical interfaces are used to construct workflows from mature scripts, not for the development of workflow steps. Consequently, users are required to develop workflow steps in other environments before wrapping them to a workflow system.
SoS Notebook is a multi-language IDE for the SoS workflow engine that provides an interactive environment for all stages of workflow development. As demonstrated in blog posts SoS Notebook: One Notebook, Multiple Kernels and Combined power of SoS and SoS Notebook, SoS Notebook allows you to:
Develop scripts in different languages using multiple kernels, with added features such as string interpolation (magic %expand) and line-by-line execution.
Convert scripts to SoS “actions” to be executed as (scratch) workflow steps. The “actions” are Python functions and can be debugged in the SoS kernel
“Chain” the scratch workflow steps into SoS workflows by adding
input,output,dependsstatements and section headers.
The SoS environment makes it easy to convert scripts developed in interactive data analysis to a workflow which removes the biggest obstacle in the utilization of workflows in daily computational researach.
Workflow system that can be comprehended in minutes
SoS is extended from Python 3.6 so a SoS script is essentially a Python script with additional workflow specifications. With Python being one of the easiest scripting language to learn and one of the most widely used scripting languages, SoS has a very smooth learning curve. As a matter of fact, SoS adds very few additional syntaxes to Python and the follow table summarizes them all:
| Syntax | Example | Usage |
|---|---|---|
| Script format of function call | sh: |
Calling a Python function with multi-line script as first parameter |
| Section header | [step_10] |
Define workflow steps |
| Parameter definition | parameter: cutoff=5 |
Define command line argument |
| Step input, output, and depends | input: “a.txt” |
Define input, output, and dependent targets of steps |
| Task | task: walltime=‘24h’ |
Define external tasks |
As shown in the following figure, a SoS workflow can be created as verbatim inclusion of scripts in different languages. The script can be annotated with additional workflow syntax to make use of more and more workflow features such as runtime signature and external execution of tasks, but the workflow remains readable because the key parts of the workflow, namely the included scripts, remain largely untouched.
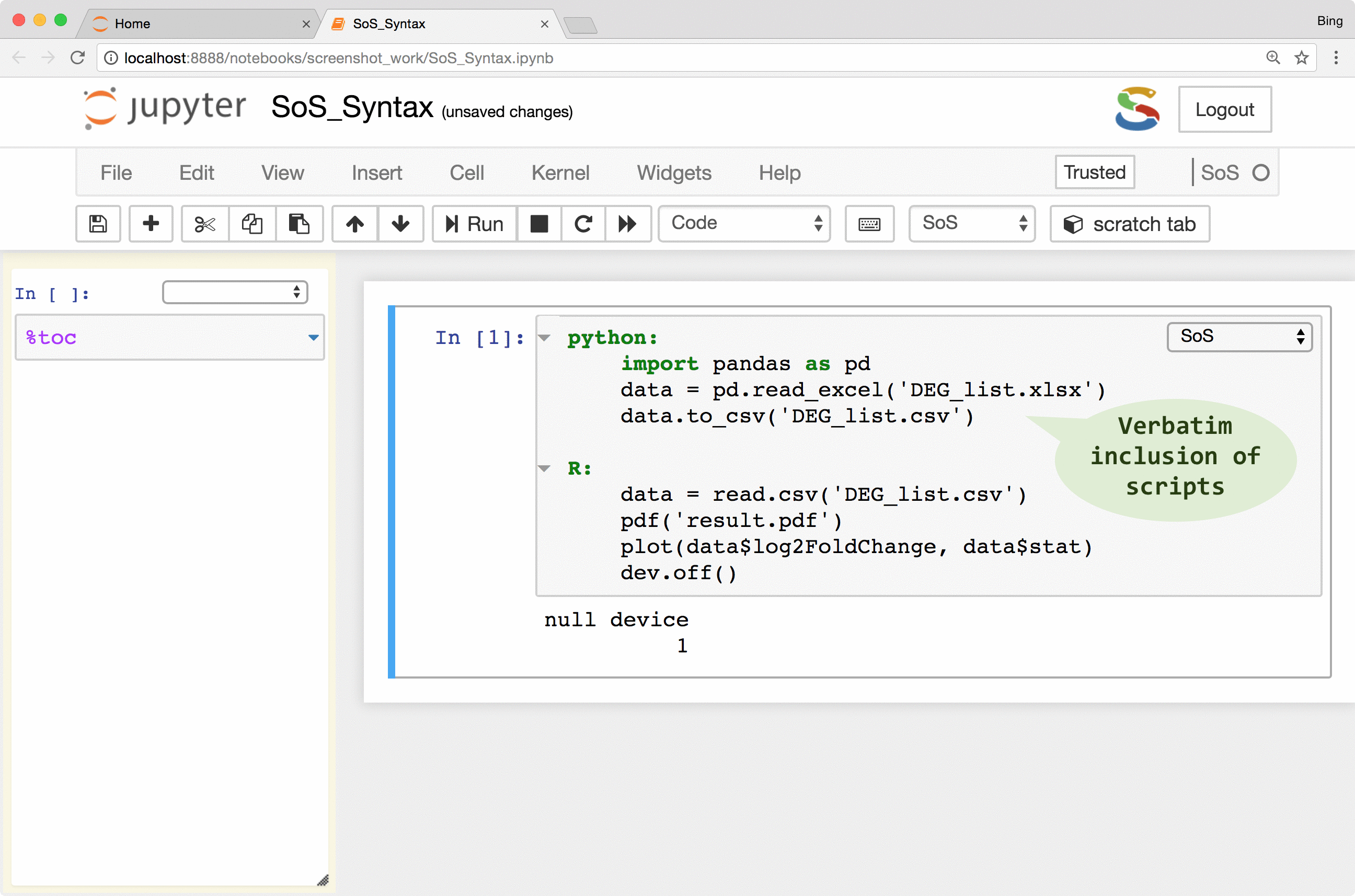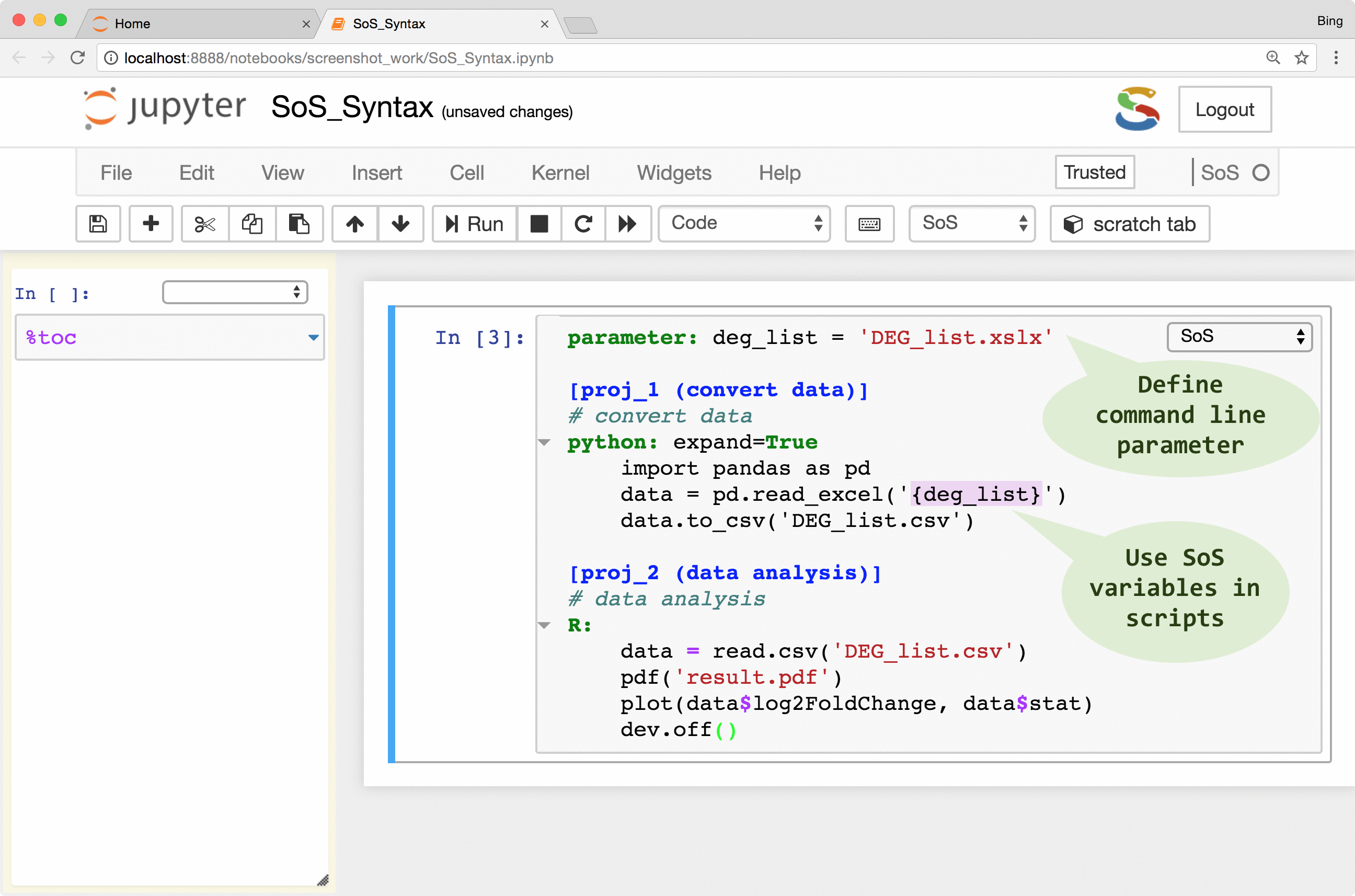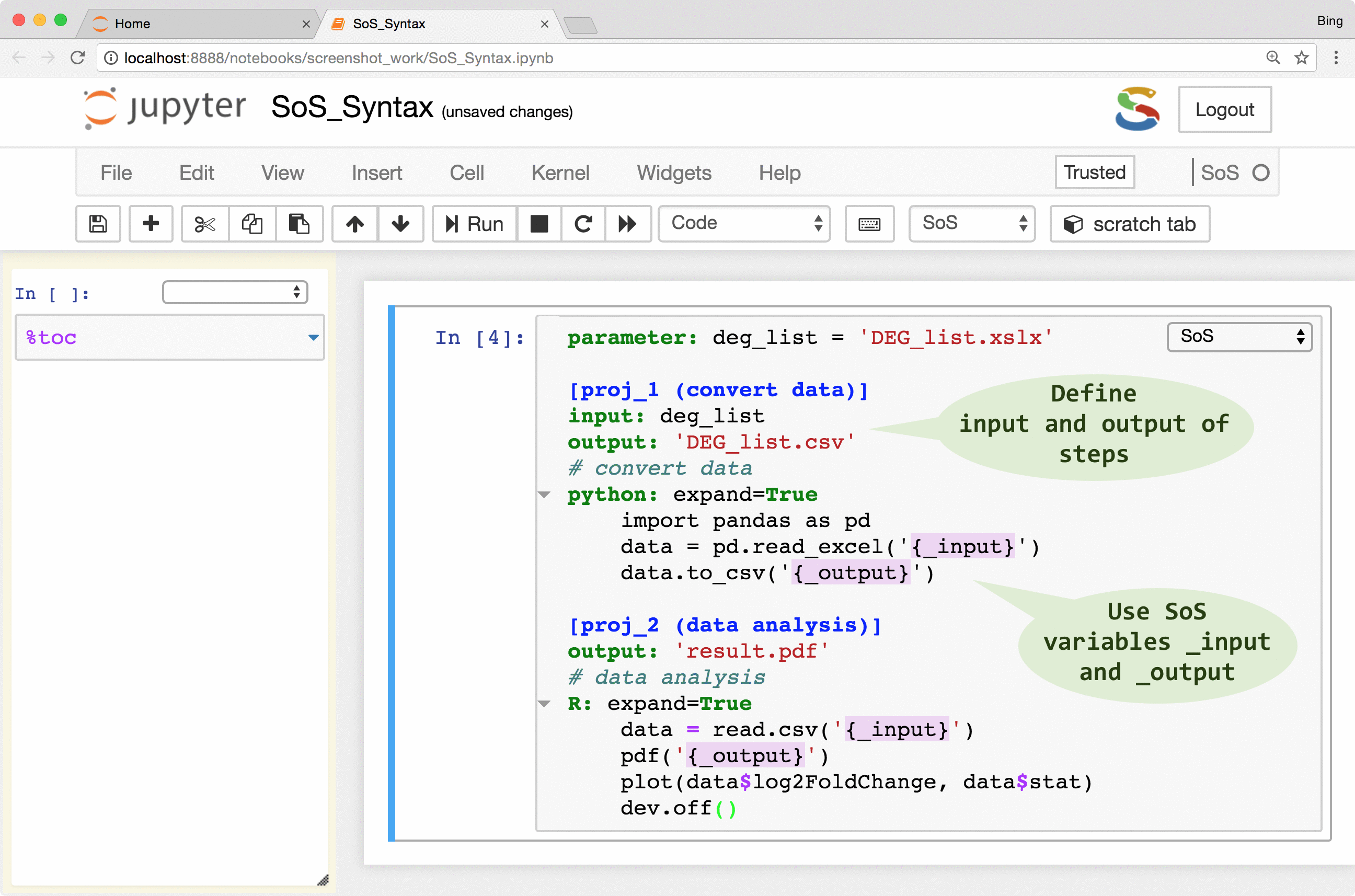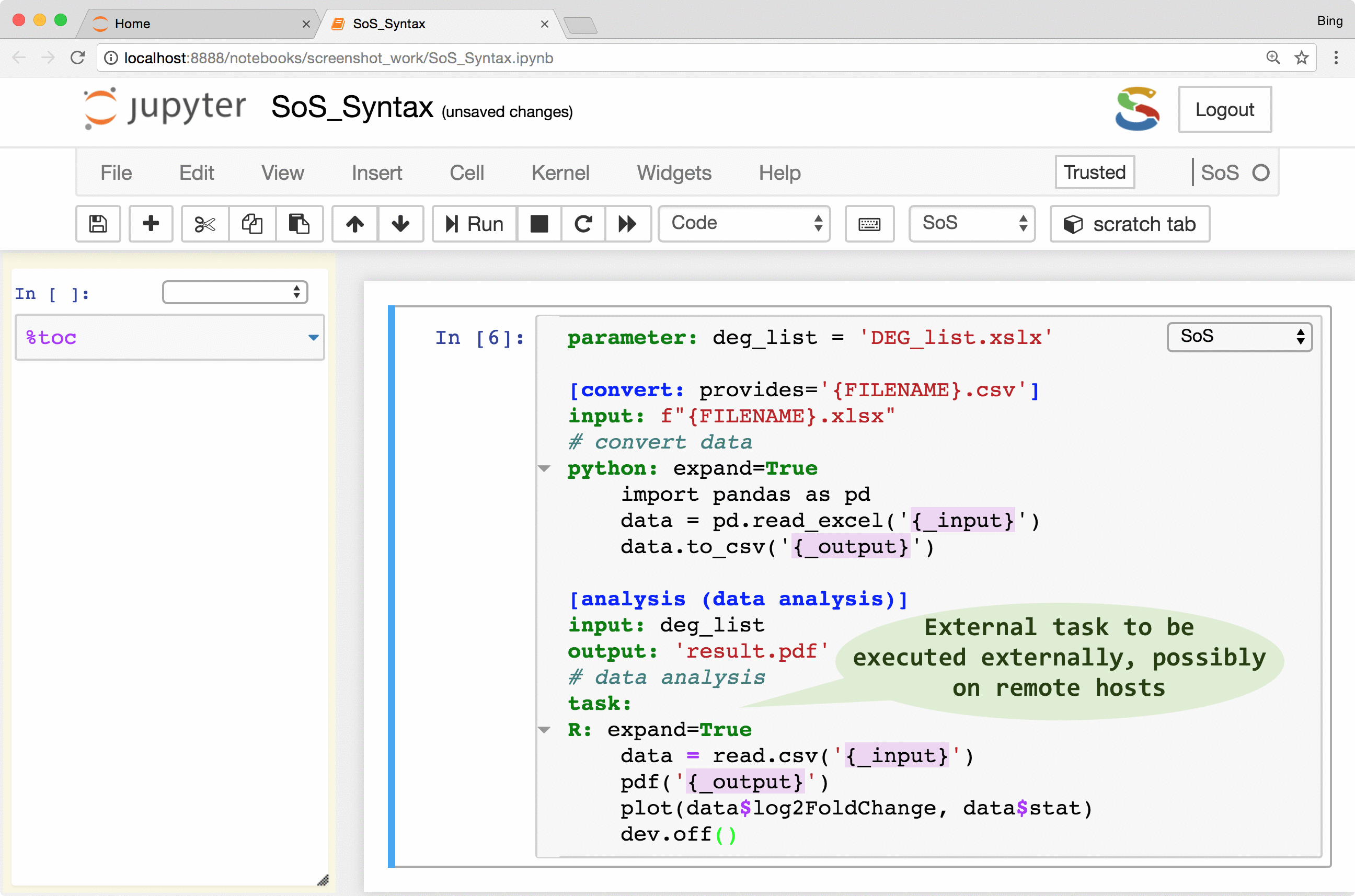
The following videos describes these steps in detail:
Benefits of using SoS for daily computational research
The moment you convert a script to a SoS workflow step, you have a working SoS workflow that can benefit from many of SoS’ execution features.
Runtime signature
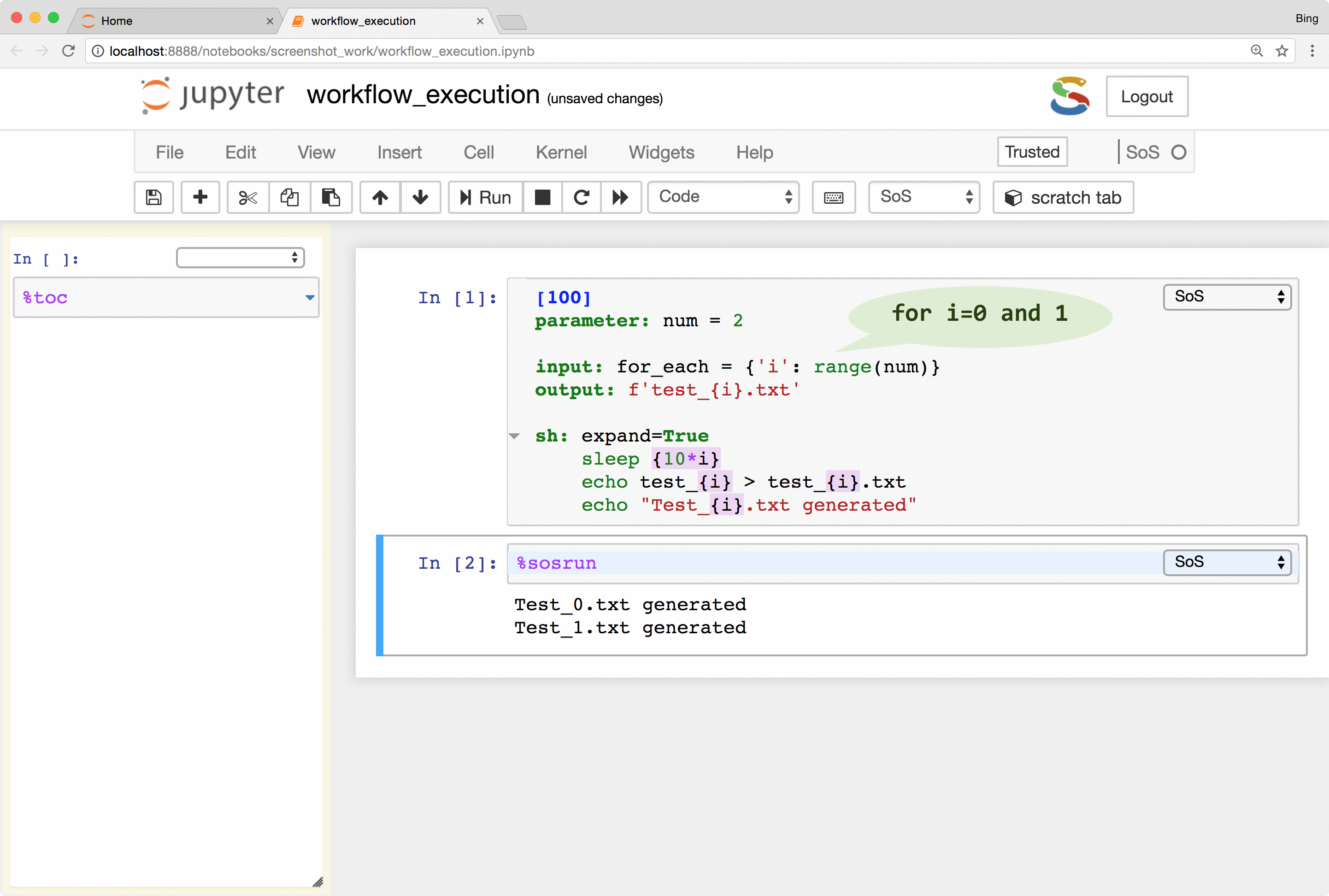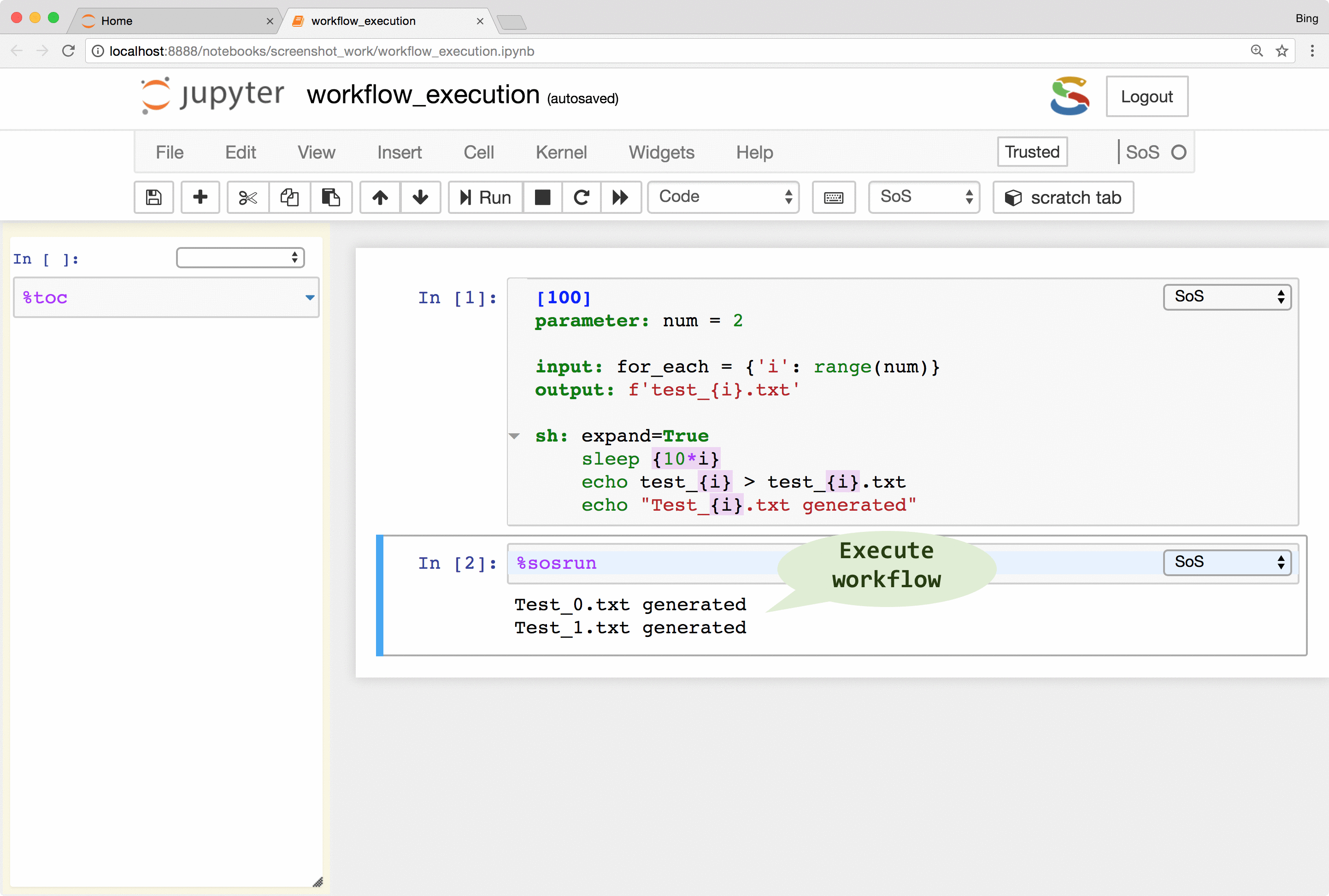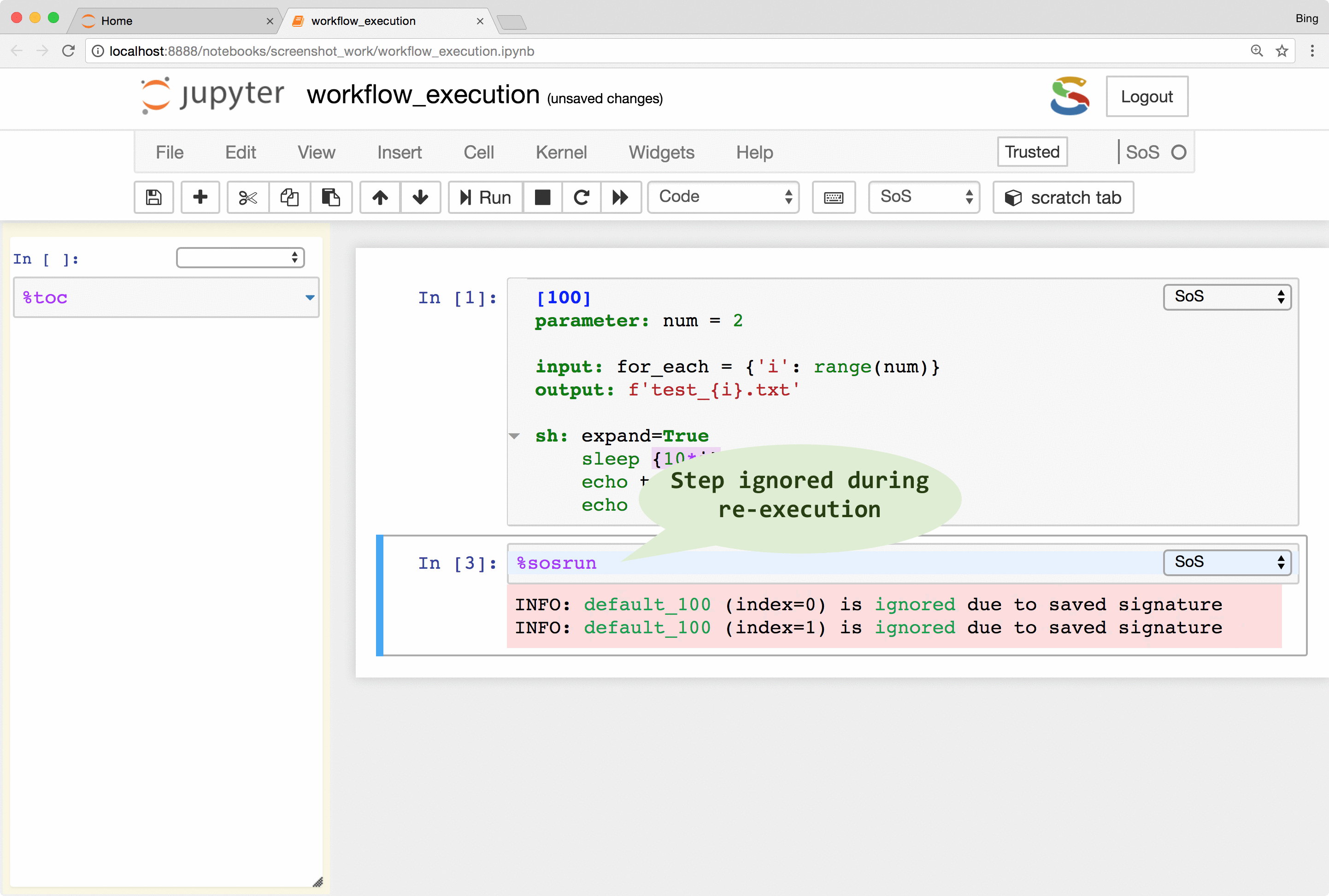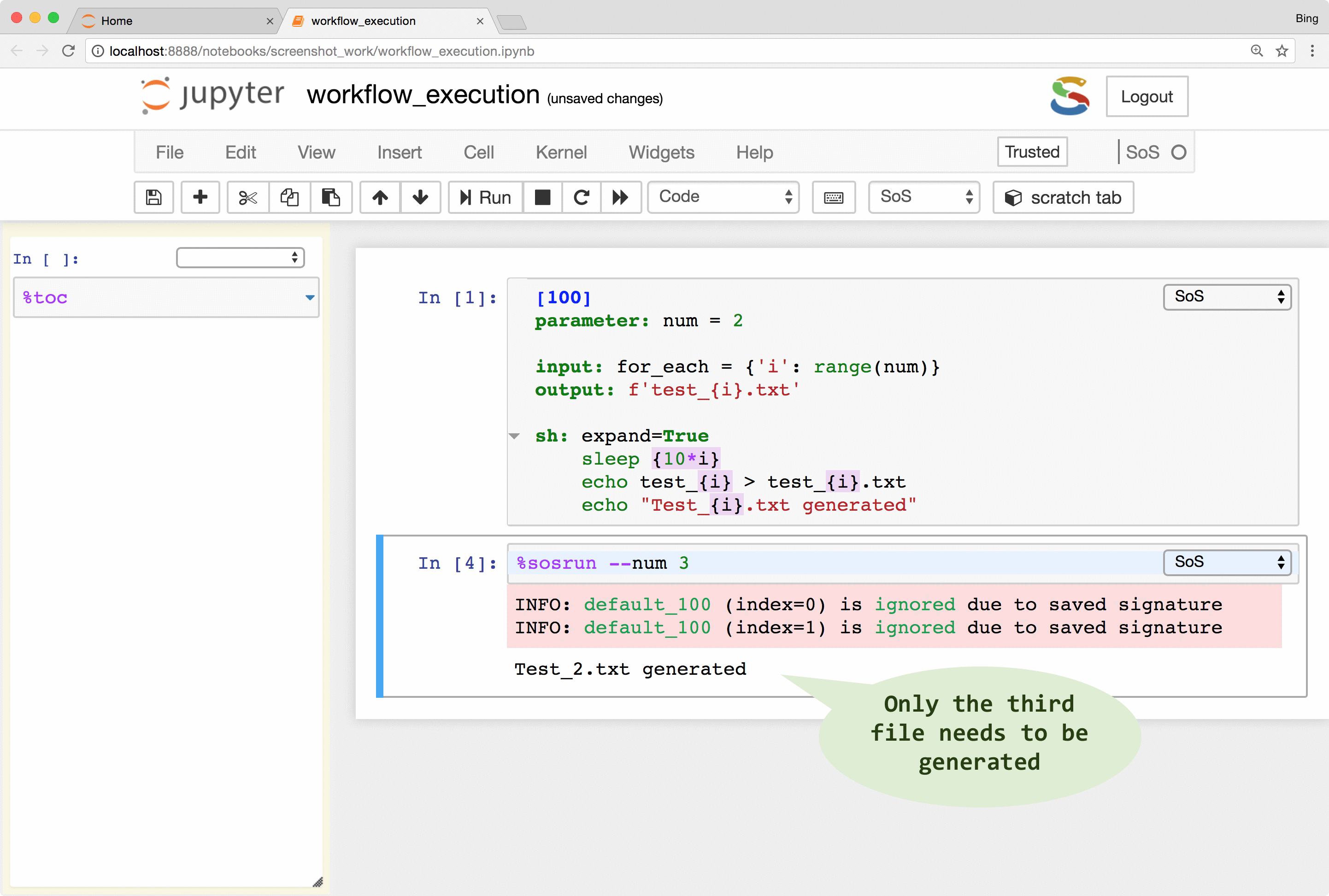
SoS keeps track of runtime signatures so it will ignore a step if it has been executed before. The signatures consist of input, output, dependent targets, statements and their environments (e.g. used global variables) and a step would be re-executed if any of these items has changed. The signatures are independent of step names and workflows so a step would be ignored even if it belongs to multiple workflows.
What this means is that you can continuously revise and execute a workflow without wasting time on the execution of steps that have been executed before, and without worrying about the re-execution of steps that should be re-executed because of changed input or parameters.
Remote execution
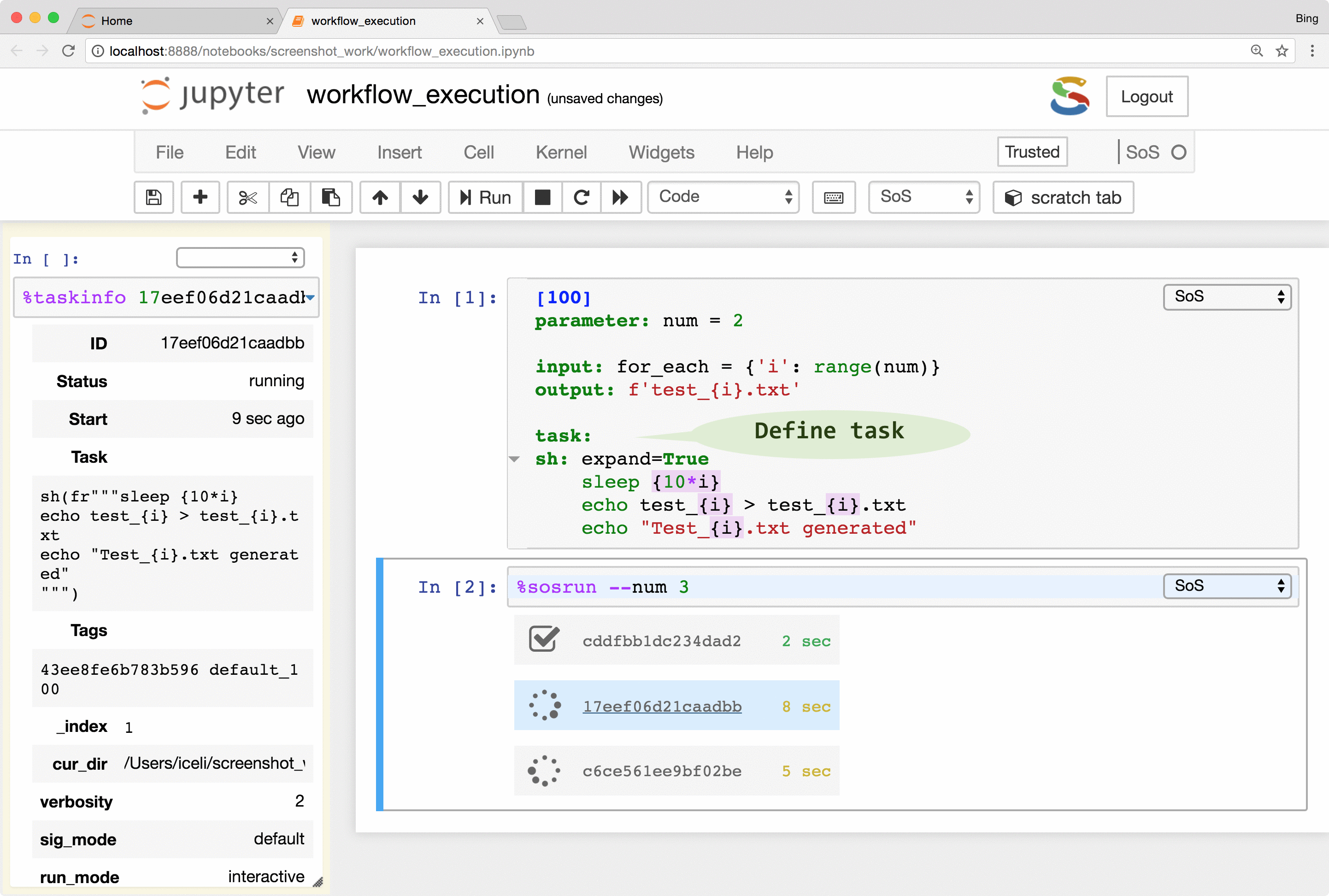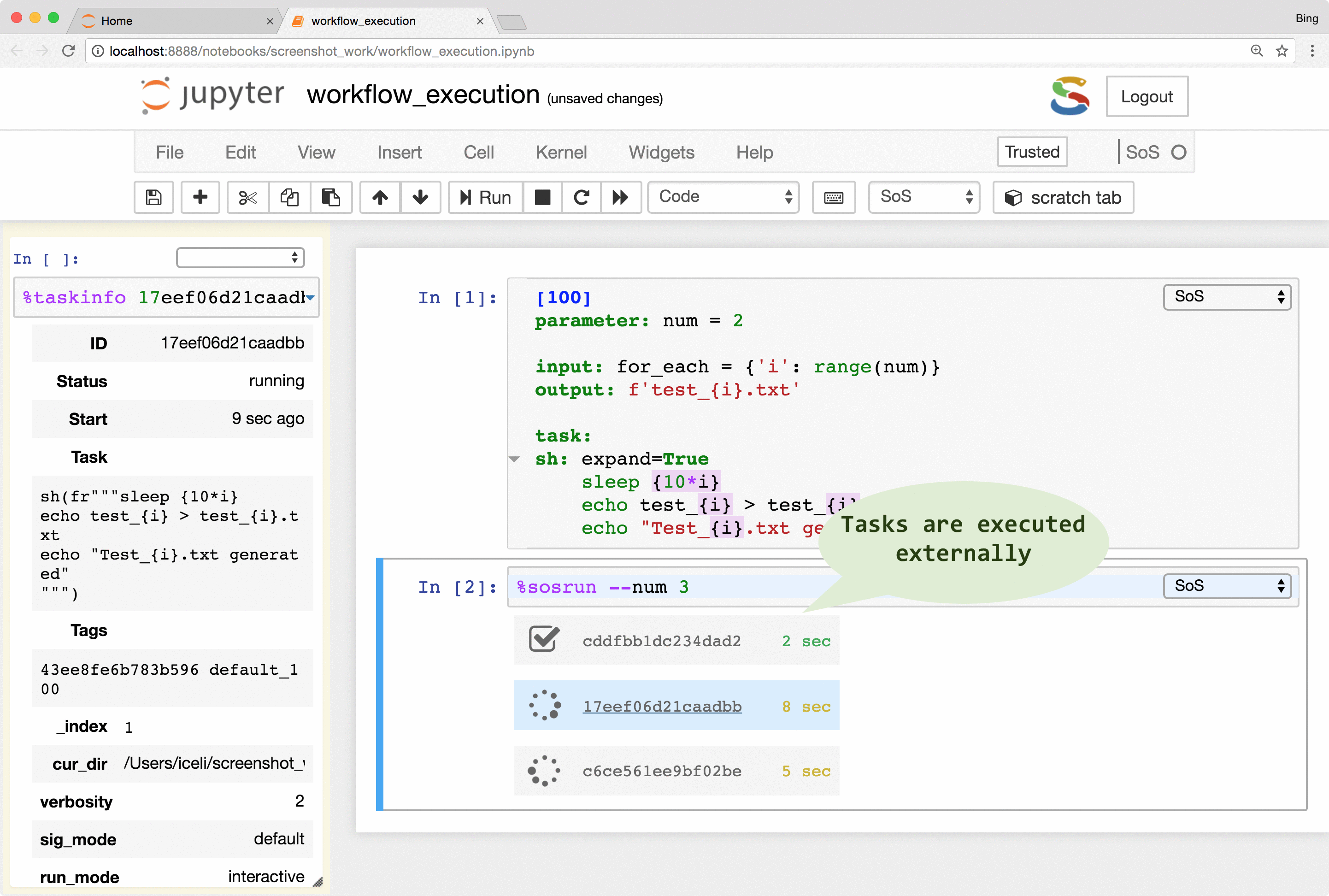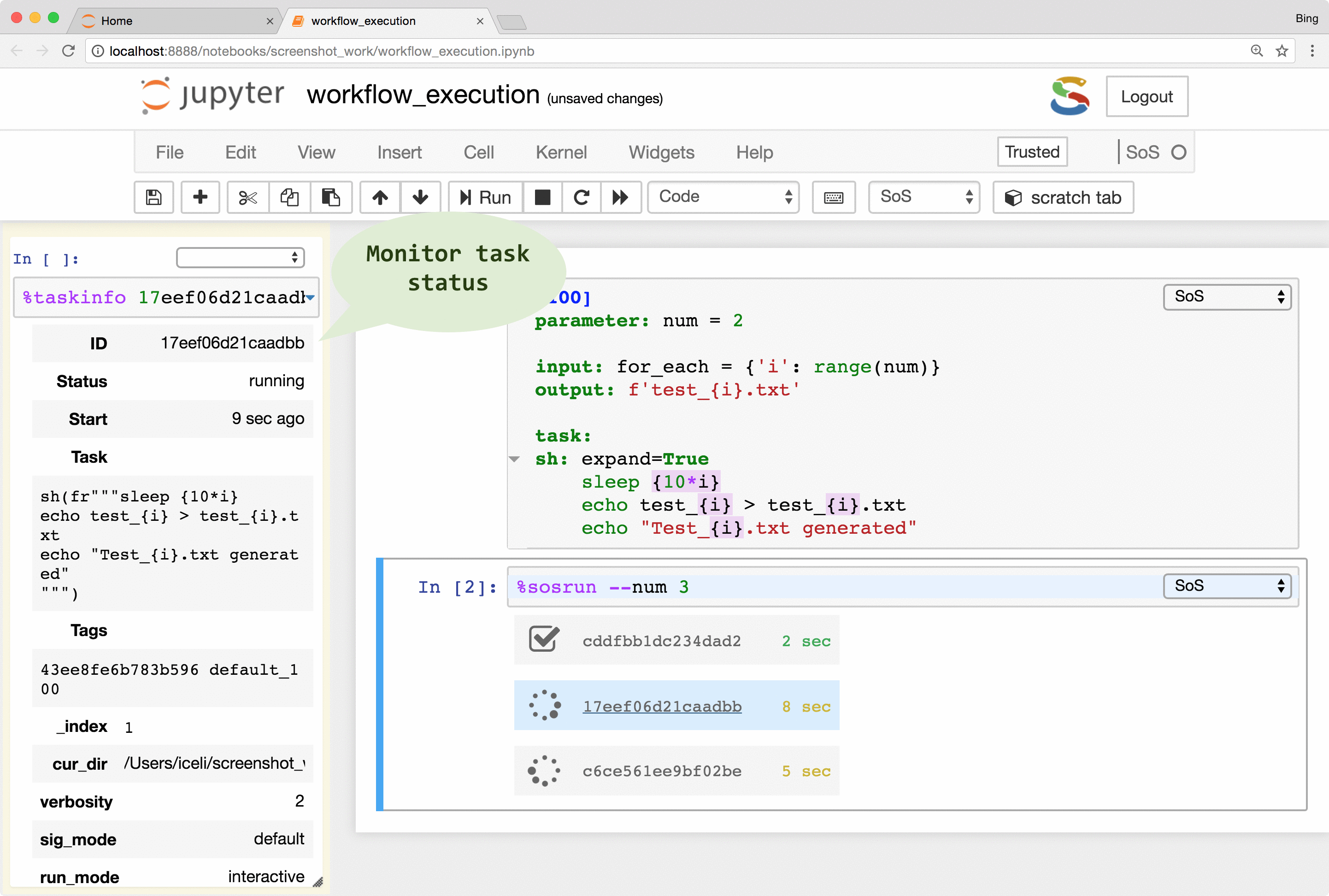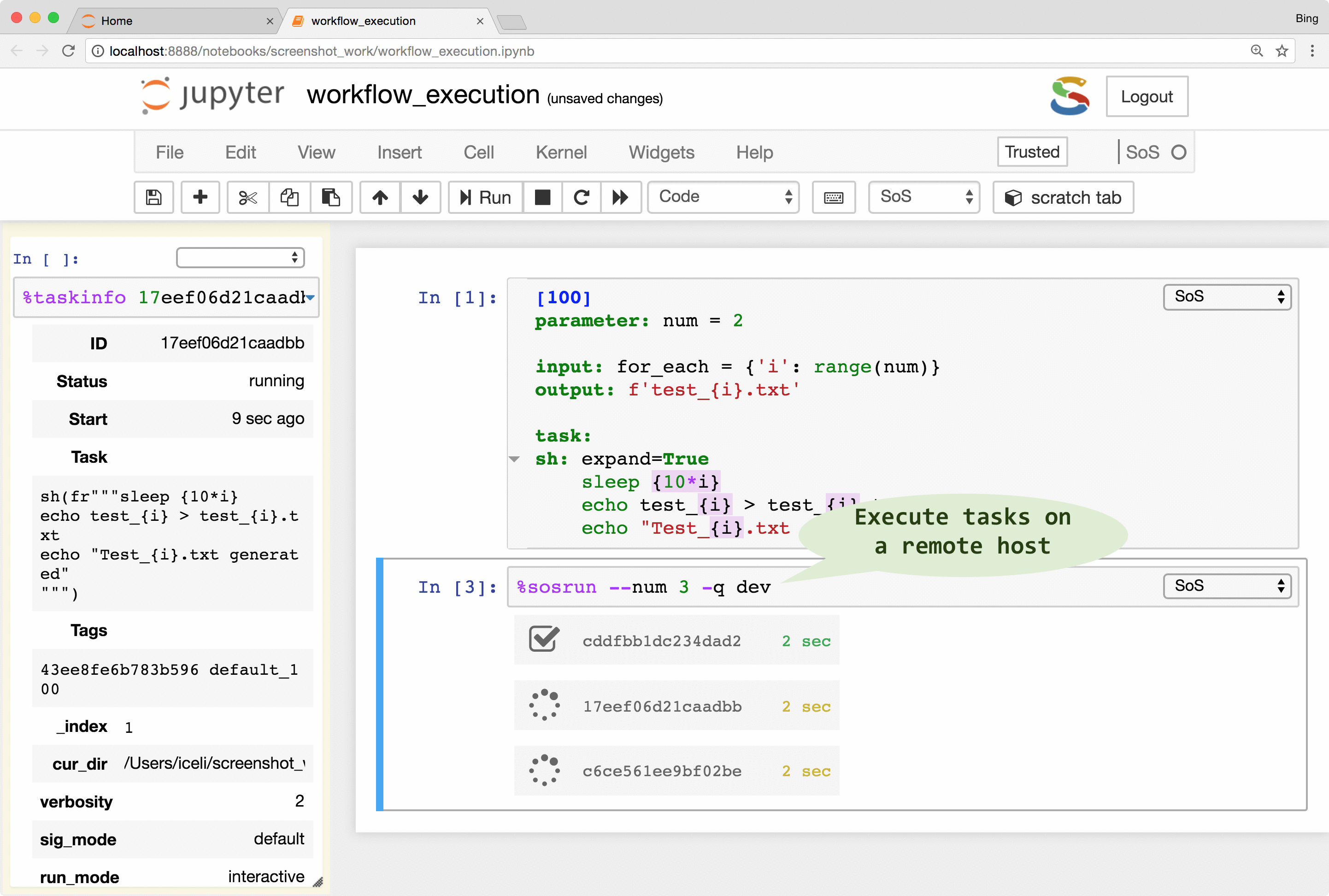
SoS allows the creation of tasks from part of a workflow step. The tasks are execution units that are independent of workflows and even file systems so they can be executed locally or remotely in parallel (subject to number of concurrent running tasks). With properly configured remote hosts, the same tasks could be executed locally or on a remote host as separate processes, or be submitted to a PBS (Moab/torque, Slurm, IBM LSF) system, or task queue systems such as RQ. The remote system does not have to share the same file system as the local host because SoS automatically synchronizes input files, updates scripts, submits and monitors jobs, and collects results. It even provides a job monitor system that is independent of the underlying job systems.
The remote execution feature gives users easy access to all available computing resources. It frees users from writing platform-specific codes for the submission and monitoring of jobs and to synchronize input and result files across systems. It also helps the readability and reproducibility of analysis because both local and remote scripts are kept in the same notebook, without platform-specific job management code.
Powerful workflow systems
SoS supports both forward-style procedure-oriented and makefile-style outcome-oriented workflows. The forward-style workflows are specified as numerically ordered steps that will be executed sequentially (logically speaking), and makefile-style workflows are specified as steps that provides targets that are used by others. SoS also supports mixed style workflows (forward-style workflow with dependencies satisfied by makefile-style steps) and nested workflows (execution of subworkflows as function calls) which allows the creation of very complex workflows that consists of hundreds of steps. The following figure illustrates the DAG of a RNA Seq data analysis workflow.
Readable workflows that are easy to report, share and reproduce
SoS Notebook provides an ideal format for the distribution of SoS workflows and the reporting and sharing of bioinformatic data analysis because a SoS notebook can include all details of a data analytic workflow, including descriptions of data and analytic steps (in rich text format with tables and figures), scripts in multiple languages, workflows, and outputs of analysis. The all-in-one nature of the notebook format also makes it easy to reproduce prior analyses if needed.
The following is an example workflow with steps in Python, R, and bash. It looks just like other online tutorials you have seen but its notebook version contains a complete workflow and is readily applicable to your data. The readability of SoS workflows distinguishes SoS from other workflow systems and makes it a great tool for the sharing of data processing expertise among bioinformaticians.
Summary
In summary, SoS provides an environment for both interactive data analysis and batch data crunching. It has a smooth learning curve because it is based on a widely used scripting language (Python) with few additional syntaxes. It cures pipelineitis by providing an easy transition from interactive to batch data analysis so users can enjoy features such as runtime signature and remote execution with minimal effort. At the same time, SoS provides a powerful workflow engine for the creation of complex workflows so whereas you can complete most of your projects use SoS Notebook and simple workflow features, your analysis can grow as your project grows, all without leaving your familiar SoS environment.
More details of SoS and SoS Notebook can be found at the SoS website where you can find tons of documentations, tutorials, examples, and youtube videos. Please test SoS Notebook and send your feedback and/or bug reports to our github issue tracker. If you find SoS Notebook useful, please support the project by starring the SoS and SoS Notebook github projects, or spreading the word with twitter.
